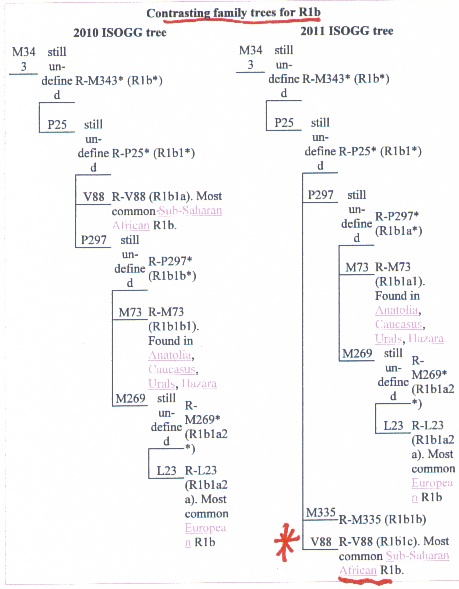
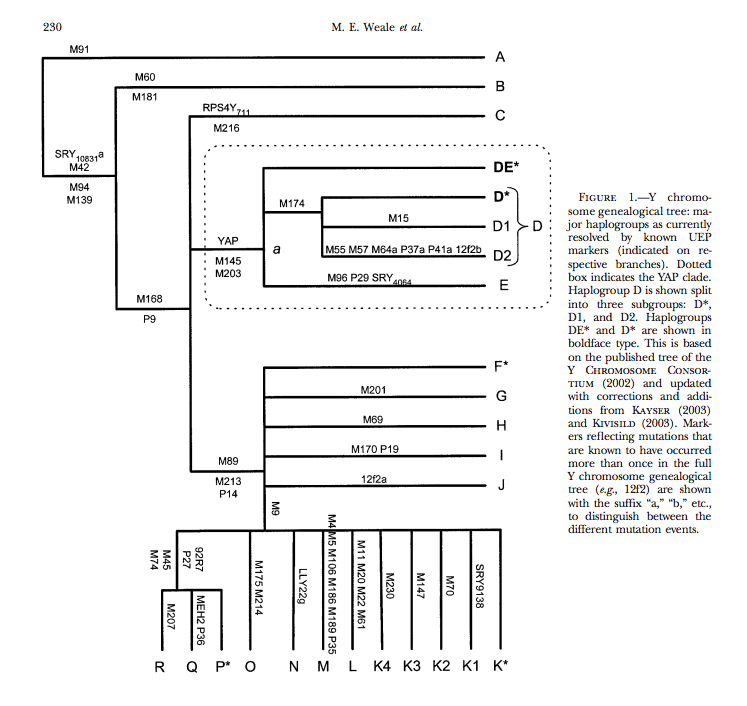
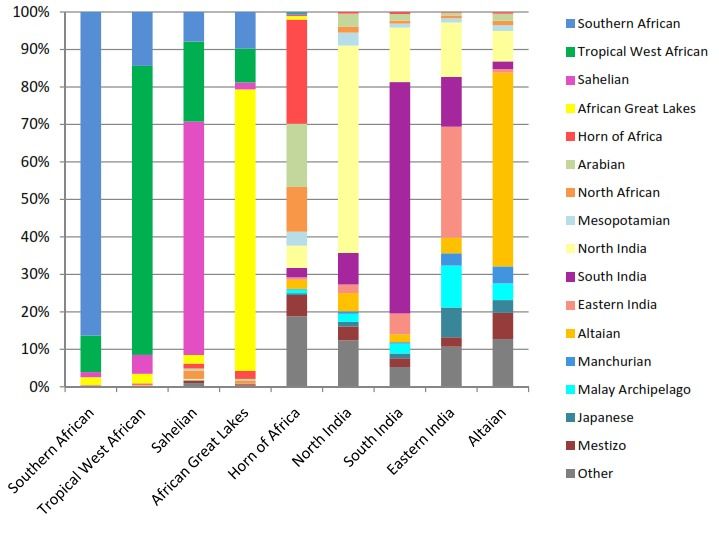
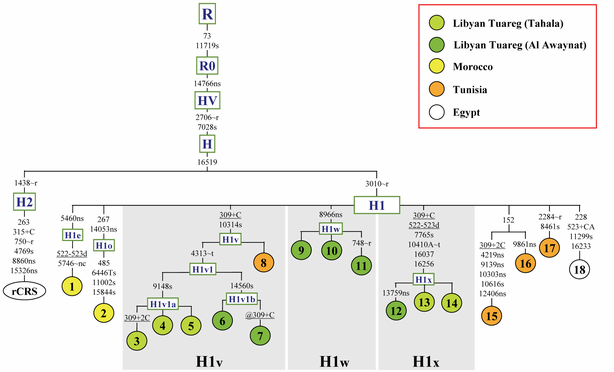
code:
Bowcock CS 1987 Study of 47 DNA markers in five populations Gene Geogr. 1(1) 47-64. Apr 1987 1987 Apr;1(1):47-64.
Bowcock cs 1991 Human Evol DNA polymorphisms RFLPs Proc. Nati. Acad. Sci. USA Vol. 88, pp. 839-843, February 1991 Evolution
Bowcock cs 1991 Study of an additional 58 DNA markers Gene Geogr. 5(3) 151-73. Dec 1991 1991 Dec;5(3):151-73.
Bowcock Kidd CS 1994 microsatellites Nature Vol. 368 pp. 455-457, March 1994 Letters to Nature
Mountain CS 1994 nDNA RFLPs Proc. Nati. Acad. Sci. USA Vol. 91, pp. 6515-6519, July 1994 Evolution
Lin CS 1994 ??? data Gene Geogr. in press in press in press 1994
Cavalli-Sforza 1994 History Geography Human Genes Book Ch. 2 89-93 1994
Cavalli-Sforza 1997 Genes Peoples Languages article Proc. Natl. Acad. Sci. USA Vol. 94, pp. 7719–7724, July 1997 Colloquium Paper
Cavalli-Sforza 2000 Genes Peoples Languages book Book 75-76 2000








![[Confused]](confused.gif)
![[Embarrassed]](redface.gif)
![[Roll Eyes]](rolleyes.gif)









![[Big Grin]](biggrin.gif)



![[Cool]](cool.gif) .
.
























![[Eek!]](eek.gif)
![[Wink]](wink.gif)
![[Smile]](smile.gif)