Originally posted by argiedude:
quote:What about India?
We know that slaves sent to the US were roughly split 60%/40% between West Africa and Central Africa (Angola/Congo).
quote:Eritrea won't differ that much from highland Ethiopians and eastern Sudan, i doubt there are any high frequencies of E3a in Eritrea since its inhabitated by only Nilosaharan, Semetic and Cushitic speakers.
argiedude:
3) The countries for which we are desperately needing y-dna data are: Sudan, Congo, Angola, Mozambique, Eritrea, Liberia.
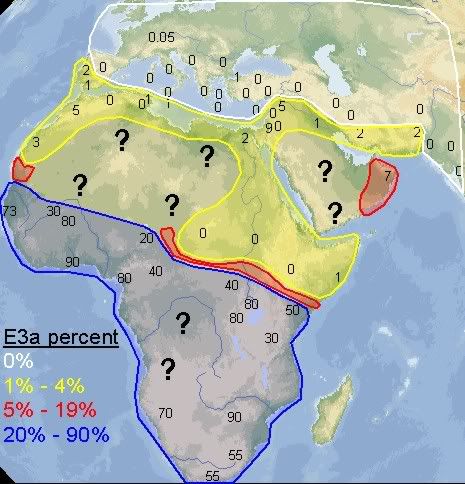
quote:E3a lineages in Madagascar reflect part of the African component in a population, where the African component is no less recent than the non-African component. The African component is no less significant either, and E3a appears to be quite substantial in this region. As for the issue of frequency distribution across the continent, well, the question put forth to potential readers, is what they thought about the map. As such, I'm of the opinion that, for the sake of precision, the presence of E3a across the continent is worth noting, whether it is sizeable or not in frequency.
Originally posted by argiedude:
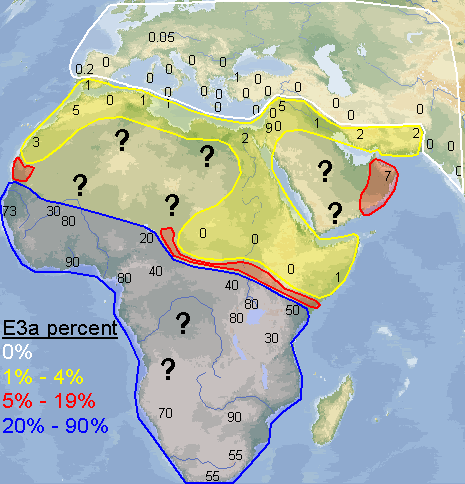
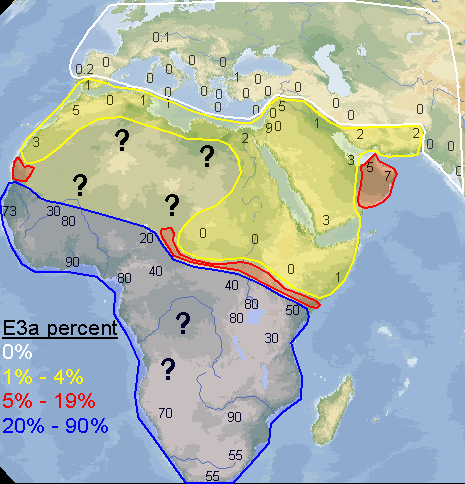
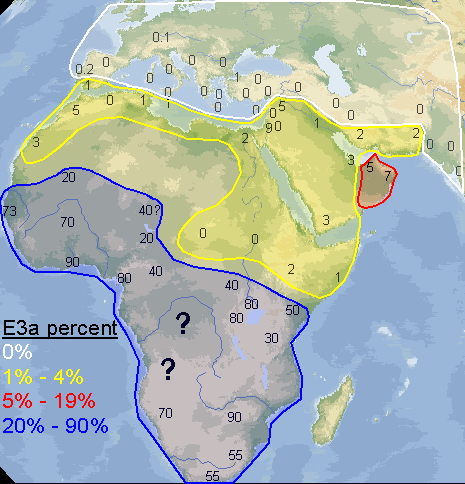
Yes, I made the map myself. I didn't include northern Africa because it's clear it doesn't have a significant percentage of E3a, and I did the map to see exactly how frequent E3a is in the places where it's supposed to be very frequent. I wanted to see if there was some sort of pattern, something interesting that would appear. That's also why I didn't include Madagascar, it's E3a would simply reflect very recent population movements and not be able to tell much about how E3a originally diffused throughout Africa. I did get a couple of useful thoughts from doing the map.

quote:Let me state that you brought up the above, not me or anyone else.
We know that slaves sent to the US were roughly split 60%/40% between West Africa and Central Africa (Angola/Congo).
quote:What's wrong with you? There's no reason to be aggressive. This unwarranted hostility is what's bringing this forum down in quality. His comments are perfectly reasonable. Are you going to object to the geographic divisions East, West, Central, and Southern Africa now, just because they are ill-defined? He's simply using it as a way to define two separate clusters of AA ancestry: the Guinean coast (Senegal to Nigeria) and the trade from Angola and Congo, which do represent distinct ancestries. Indian (as in from India) admixture in AAs is very minimal and not at issue here, since we're discussing E3a, which is prominent in Africans and rare in Indians.
Originally posted by One_and_Done:
argiedude wrote:
quote:Let me state that you brought up the above, not me or anyone else.
We know that slaves sent to the US were roughly split 60%/40% between West Africa and Central Africa (Angola/Congo).
So now are you going to answer this question. What about India? Because there were Indians (from India) brought to America. As well as others. There is even a thread about it.
So how can there be this "60%/40% between West Africa and Central Africa (Angola/Congo)" ? Also while you're at it define what countries make up "West Africa".
Apparently you either don't know what you are talking about or you are a bald faced liar with a severe dogmatic psychosis.
quote:I'm not getting agressive. I asked the guy a simple question about the information that he placed in this post. It seems that you're the one getting all "bug-eyed" and aggresive here.
What's wrong with you? There's no reason to be aggressive. This unwarranted hostility is what's bringing this forum down in quality. His comments are perfectly reasonable. Are you going to object to the geographic divisions East, West, Central, and Southern Africa now, just because they are ill-defined? He's simply using it as a way to define two separate clusters of AA ancestry: the Guinean coast (Senegal to Nigeria) and the trade from Angola and Congo, which do represent distinct ancestries. Indian (as in from India) admixture in AAs is very minimal and not at issue here, since we're discussing E3a, which is prominent in Africans and rare in Indians.
quote:So does this mean it is unkown if E3a is found in countries like Chad, Niger, and Mauritania?? I thought it was even present in Egypt?!
Originally posted by argiedude:

quote:I didn't find any references to y-dna studies for those 3 countries you mentioned, but at the same time, even if I had info for them, their populations are heavily concentrated in the south of their states, so that big blank spot in the Sahara region wouldn't change much.
Originally posted by Djehuti:
quote:So does this mean it is unkown if E3a is found in countries like Chad, Niger, and Mauritania?? I thought it was even present in Egypt?!
Originally posted by argiedude:
quote:And you call me aggressive? Do you even know the meaning of that word? Colorless green ideas sleep furiously!
Originally posted by One_and_Done:
Yom foraged:
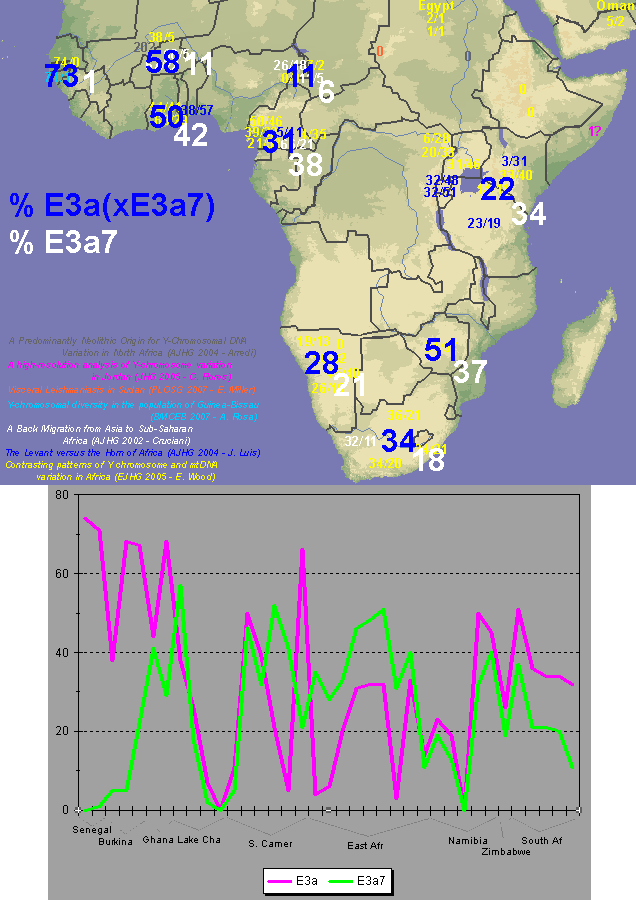
quote:No, you are the idiot here. First of all, you don't know where I was educated. Secondly, 40% and 60% are both approximations corresponding to African contributions to the AA genepool. Obviously some 17% is Caucasian that wasn't originally present, and maybe 1% is Native American that wasn't found in the first enslaved Africans in North America. You can still take those non-African influences into account, and look at the breakdown between E3a7 and E3a*(xE3a7) to get an approximation or vague idea of its frequency in Central Africa, since you know it's about 2:1 (of E3a lineages, of course) in West Africa. Unfortunately, West Africa isn't homogenous and Senegal is an extreme case, as well as a point of departure for many slaves (less so a point of origin but that too), so the formulated picture is very rough.
I'm not getting agressive. I asked the guy a simple question about the information that he placed in this post. It seems that you're the one getting all "bug-eyed" and aggresive here.
Do you not know how to add? What is 60% + 40%?
Do you now this basic calculation and what it means? Please don't tell me that the school systems in your country are that bad.
That is something that even the most low achieving student in middle school already knows.
60% from "West Africa", 40% from "Central Africa" = 100%. Meaning nobody else came from anyplace else.
Which is wrong because we know that people were brought over from India as well as other places.
So he's posting misinformation either on purpose because of some fantasy or some other form of psychotic dogma.
quote:You made the claim, back it up. I have studies on the composition of African Americans, and not a one mentions Indian admixture from India. That's not to say that Indians never mixed with African Americans, but they didn't do so often enough and in large enough numbers to significantly impact their gene pool.
And as far as all of this Indian admixture is concerned I take what you say with a grain pepper seing as that is nothing more than your opinion. As far as I'm concerned I don't know who mixed with who and that is quantified as to if there was ever any admixture.
quote:What studies are you deriving this information from?
Originally posted by Yom:
Probably not, given that it's not present in N. Sudan. IIRC, it's present at 8% in Upper Egypt. It is common among Siwis (Siwa Berbers), though, along with E3, E2, E1, etc. E3b is actually not commonly found in them.
quote:I could really use that information (north Sudan, south Egypt and Siwas) to better this map. If you can remember where it's from, I'd be very thankful.
Originally posted by Yom:
it's not present in N. Sudan. IIRC, it's present at 8% in Upper Egypt. It is common among Siwis (Siwa Berbers), though
quote:Do you see a claim here other than I don't know who mixed with who if it even occured?
You made the claim, back it up. I have studies on the composition of African Americans, and not a one mentions Indian admixture from India. That's not to say that Indians never mixed with African Americans, but they didn't do so often enough and in large enough numbers to significantly impact their gene pool.
quote:The scientists.........Okay, got it now?
There are no such clowns here for the most part, quite the contrary, so what are you talking about?
quote:BTW -
Originally posted by reserved:
You people have way to much time on your hands. What the **** is an E3a?
quote:Am I reading this map correctly? Is ther 9% E3a in Palestine? If so, is there a historical explaination for this Sub-Saharan gene in Israel? We are cetainly not talking Natufian era.
Originally posted by argiedude:
Update after a couple of new E3a data came out in the last few months.
2 E3a samples (same haplotype) out of 194 samples were found in the Italian Tyrol Alps (German speaking region bordering with Austria).
The very recent study about y-dna in Oman, Qatar, and U.A.E.
quote:I don't have no clue what you're talking about. I take it an E3a is an African? What is the origin of the word?
Originally posted by xyyman:
Here is another fuhl?
quote:BTW -
Originally posted by reserved:
You people have way to much time on your hands. What the **** is an E3a?
the E3a in Italy/Germany. Are you implying it is ancient? or the new trend? Having been to Germany several times in the last five years. Seem like the African bros are a hot commodity. That's why I am asking.
quote:Though not entirely accurate and perhaps a bit misleading but this URL should help you understand what this thread is about:
Originally posted by reserved:
quote:I don't have no clue what you're talking about. I take it an E3a is an African? What is the origin of the word?
Originally posted by xyyman:
Here is another fuhl?
quote:BTW -
Originally posted by reserved:
You people have way to much time on your hands. What the **** is an E3a?
the E3a in Italy/Germany. Are you implying it is ancient? or the new trend? Having been to Germany several times in the last five years. Seem like the African bros are a hot commodity. That's why I am asking.
quote:Maybe, the slave trade sounds like a more reasonable explanation for most of it, though. There are still "Black Palestinians" living in Palestine whose ancestors have been there for centuries.
Originally posted by osiriun:
quote:Am I reading this map correctly? Is ther 9% E3a in Palestine? If so, is there a historical explaination for this Sub-Saharan gene in Israel? We are cetainly not talking Natufian era.
Originally posted by argiedude:
Update after a couple of new E3a data came out in the last few months.
2 E3a samples (same haplotype) out of 194 samples were found in the Italian Tyrol Alps (German speaking region bordering with Austria).
The very recent study about y-dna in Oman, Qatar, and U.A.E.
quote:Very probable, after all this lineage wasn't born yesterday, nor were its bearers ever immobile as far as I know.
Originally posted by osiriun:
Though not entirely accurate and perhaps a bit misleading but this URL should help you understand what this thread is about:
https://www3.nationalgeographic.com/genographic/
E3a has an interesting history. Though it certainly left the Nile Valley well before the Dynastic period,
quote:Many Black Palestinians result from the fact that West Africans after making the hajj to Mecca, made pilgrimage to sites in Jerusalem relating to Muhammad and Islam. These hajis remained in Palestine and represent some of the Black Palestinians.
Maybe, the slave trade sounds like a more reasonable explanation for most of it, though. There are still "Black Palestinians" living in Palestine whose ancestors have been there for centuries.
quote:With what would the blackness of Levantine populations be synonymous? Do you know at what period the Natufians left Africa and went into the Levant?
Originally posted by alTakruri:
Just judging from whitedude's mapping:
Slave trade? Preposterous!
9% in Israel/Palestine but
not exceeding 3% in Arabia
or 5% in Morocco?
Highly unlikely.
As far as "black Palestinians,"
they were proceeded by other
presumed blacks; Natufians and
Canaanites and Phillistines and
Judahites (the last three also
having non-black elements too).
Nor is the "blackness" of any of
these Levantines synonymous with
E3a.
quote:I was wrong, mtdna L in Yemen is actually 47% (from a sample of 115).
Originally posted by argiedude:
But the black female ancestry is lower than that: mtdna L amongst Arabs is much less than 25%, reaching maybe 15% at the most (in Yemen?).
quote:I have actually heard arguments that sickle cell anemia is not of African origins.
Originally posted by Mystery Solver:
This notion that E3a lineages outside of Africa equates to "E3a=Slavery" is essentially intellectual bankruptcy. This is what some dogmatic characters do even with certain mtDNA lineages, especially when it is predominantly found in West and Central Africa - attribute it to slavery. Take L1b [predominantly detected in West Africa] for instance, which shatters such dogma-infested rationale:
Mitochondrial DNA sequences and restriction fragment polymorphisms were retrieved from three Islamic 12th-13th century samples of 71 bones and teeth (with >85% efficiency) from Madinat Baguh (today called Priego de Cordoba, Spain). Compared with 108 saliva samples from the present population of the same area, the medieval samples show a higher proportion of sub-Saharan African lineages that can only partially be attributed to the historic Muslim occupation. In fact, the unique sharing of transition 16175, in L1b lineages, with Europeans, instead of Africans, suggests a more ancient arrival to Europe from Africa - Casas et al.
On the same token, had HbS not been detected in Old Kingdom royal mummies, there will be clearly dogmatic personalities out there trumpeting the idea that it spread much more recently through slave trade. Even today, the HbS type found in Egypt, is of the Benin haplotype, which would have likely originated in populations now living in around Niger River Valley regions. Could the sickle cell trait have arrived in the Nile Valley some time before the Dynastic period? The indicators suggest so, and it could have arrived with populations emanating from the Saharan belt, which is another primary source for proto/pre-dynastic natives. E3a which is undoubtedly found in Egypt, including the lower Egypt, could have been amongst the various lineages that made their way into the region at various points in time. The African Horn has historically been an avenue for bringing slaves into parts of the so-called "SW Asia", and yet for the most part, little E3a is detected here; what notable detection is found here, is largely concentrated in the southern limits of the region.
quote:Here you go - supposedly 4 independent forms of Sickle cell anemia (3 in African and 1 in Arabia). Sicle cell anemia might not be the best approach to tracing recent African migrations. E3b and E3a seems to be far less controversial.
Originally posted by Mystery Solver:
^HbS in Egypt is of what non-African derivative? In fact, give me a list of HbS on either side of the Mediterranean sea, that is not of the African derivative.
quote:You must not have read the questions, and so, I repeat:
Originally posted by osiriun:
quote:Here you go - supposedly 4 independent forms of Sickle cell anemia (3 in African and 1 in Arabia). Sicle cell anemia might not be the best approach to tracing recent African migrations. E3b and E3a seems to be far less controversial.
Originally posted by Mystery Solver:
^HbS in Egypt is of what non-African derivative? In fact, give me a list of HbS on either side of the Mediterranean sea, that is not of the African derivative.
Desai, D. V.; Hiren Dhanani (2004). "Sickle Cell Disease: History And Origin". The Internet Journal of Hematology 1 (2). ISSN 1540-2649 [/QB]
quote:Well then, what is stopping you from producing the requested list, as well as HbS chromosomes found in Egypt?
Originally posted by osiriun:
Essentially what is on either side of the Mediterranean sea may not be of African origin.
quote:Correct. 3 of the 4 sickle cell forms are of African origin with only the Arab-Indian form being the only Eurasian one.
Originally posted by osiriun:
Here you go - supposedly 4 independent forms of Sickle cell anemia (3 in African and 1 in Arabia).

quote:Actually, analysis of sickle cell is just as valid as analysis of Y-chromosomes-- all are genetic studies.
..Sickle cell anemia might not be the best approach to tracing recent African migrations. E3b and E3a seems to be far less controversial.
quote:Thanks you for providing you own information and answering you own question. Wheww.
Originally posted by Mystery Solver:
In the meantime, as we let Osiriun collect his thoughts, here is some trivia that some of us have become all too familiar with:
Haplotypes of the beta-globin gene as prognostic factors in sickle-cell disease.
el-Hazmi MA, Warsy AS, Bashir N, Beshlawi A, Hussain IR, Temtamy S, Qubaili F.
Medical Biochemistry Department, World Health Organization Collaborating Centre for Haemoglobinopathies, Thalassaemias and Enzymopathies, College of Medicine, King Khalid University Hospital, Riyadh, Saudi Arabia.
"We collaborated with researchers from Egypt, Syrian Arab Republic and Jordan in a study of patients with sickle-cell disease from those countries, and from various parts of Saudi Arabia, in order to investigate the influence of genetics on the clinical presentation of the disease, and to attempt to determine the **origin** of the sickle-cell gene in Arabs. Our results suggest that beta-globin gene haplotypes influence the clinical presentation of sickle-cell disease, and that there are at least two major foci for the origin of the sickle-cell gene, one in the eastern part of Saudi Arabia, and the other in the populations of North Africa and the north-western part of the Arabian peninsula…The Benin haplotype was found in patients with severe disease, either as homozygous or in combination with another haplotype. The majority of Syrians and Jordanians had the Benin haplotype, and severe disease. However, one in three Syrians and one in five Jordanians had a milder disease, and the Saudi-Indian haplotype was identified. All Saudi patients from south-western and north-western areas, where the disease is generally severe, had the Benin haplotype in the homozygous or heterozygous state. Of the Saudi patients from the eastern area, where a mild form of SCD exists, only 9% had the Benin haplotype. The remainder had the Saudi-Indian haplotype, either in its homozygous or heterozygous state…Restriction endonuclease restriction sites have provided a useful insight into the normal polymorphic variations in the DNA surrounding various gene loci, where a combination of two or more polymorphic sites has led to the identification of specific haplotype patterns [13,14]. This has been of significance in the study of the regions surrounding the b-globin gene (i.e. the b-globin gene cluster), where several polymorphic sites have been identified, and population differences have been found on analysis of the haplotype pattern [9]. An interesting observation is that the sickle-cell mutation has occurred on chromosomes carrying different polymorphic sites and different b-globin gene haplotypes, and this seems to play a role in the clinical expression of SCD [9].
We compared the haplotype pattern of SCD patients from different Arabic-speaking countries. Benin haplotype was the major haplotype in all countries with a severe presentation of SCD and it was present in both the homozygous and heterozygous state. This was true for those SCD patients from south-western and north-western areas of Saudi Arabia, and for those from Egypt, Jordan and Syrian Arab Republic. On the other hand, patients from the eastern part of Saudi Arabia, who present with a significantly milder clinical picture, carried the Saudi-Indian b-globin gene haplotype either in its homozygous or heterozygous state."
Source: East Mediterr Health J. 1999 Nov;5(6):1154-8 http://www.emro.who.int/Publications/EMHJ/0506/10.htm
Article was also discussed here: http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=8;t=001404#000000
It is of no coincidence that the other African haplotypes outside of the Benin haplotype, have found their way into the Americas, regions that have been affected by the historic slave trade:
From these original foci of the HbS mutation, the gene spread along trading routes to North Africa and the Mediterranean, was transported in large populations to North and South America and the Caribbean during the slave trade, and latterly has spread to Northern Europe by immigration from the Caribbean, directly from Africa to the United Kingdom, France, Belgium, and Holland, and from Turkey to Germany. The relative prevalence of these haplotypes in the Americas reflects the different origins of their African peoples, approximately 70% of HbS associated chromosomes having the Benin haplotype, 10% Senegal and 10% Bantu. Haplotype frequencies in Jamaica are similar to the USA but the Bantu haplotype accounts for the majority of HbS associated chromosomes in Brazil.9 - Graham R. Serjeant, MD, FRCP, MRC Laboratories (Jamaica), University of the West Indies, Kingston.
...in contrast to...
The Benin haplotype accounts for HbS associated chromosomes in Sicily,4 Northern Greece,10 Southern Turkey,11 and South West Saudi Arabia,6,7 suggesting that these genes had their origin in West Africa. The Asian haplotype is rarely encountered outside its geographic origin because there have been few large population movements and Indian emigrants have been predominantly from non HbS containing populations. However, it is of interest that the Asian haplotype was first described among descendants of Indian indentured laborers in Jamaica.12 - Graham R. Serjeant, MD, FRCP, MRC Laboratories (Jamaica), University of the West Indies, Kingston.
^If this sounds familiar, it's because it has been posted here: http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=8;t=005240#000021
quote:Can we tell when it was introduced based on a mutation rate analysis? The map you provide seems to not answer my question. E3a is present at 9% in Palestine but where's the HbS? Also, seems that this simply jumped right past East Africa?
Originally posted by Djehuti:
quote:Correct. 3 of the 4 sickle cell forms are of African origin with only the Arab-Indian form being the only Eurasian one.
Originally posted by osiriun:
Here you go - supposedly 4 independent forms of Sickle cell anemia (3 in African and 1 in Arabia).
As you can see from the map above, the form found in the Mediterranean is Benin HBS and NOT of the Eurasian variety. Analysis show that it first entered Europe during pharaonic times or earlier (Neolithic).
There are at least four distinct African and one Asian chromosomal backgrounds (haplotypes) on which the sickle cell mutation has arisen. Additionally, previous data suggest that the beta(S)/Bantu haplotype is heterogeneous at the molecular level. Here, we report the presence of the (A)gamma -499 T-->A variation in sickle cell anemia chromosomes of Sicilian and North African origin bearing the beta(S)/Benin haplotype....
the Benin haplotype accounts for HbS associated chromosomes in Sicily, Northern Greece, Southern Turkey, and South West Saudi Arabia, indicating that these genes had their origin in West Africa. - Graham R. Serjeant, MD, FRCP
quote:Actually, analysis of sickle cell is just as valid as analysis of Y-chromosomes-- all are genetic studies.
..Sickle cell anemia might not be the best approach to tracing recent African migrations. E3b and E3a seems to be far less controversial.
quote:I would assume this is the case since geneticists have estimated the time period it was introduced to Europe.
Originally posted by osiriun:
Can we tell when it was introduced based on a mutation rate analysis?
quote:E3a is a Y-chromosomal haplotype. HbS is a sicklecell haplotype. Just because someone has E3a lineage, doesn't mean he carries Hbs! The map presented only shows the distribution of the sicklecell haplotypes. Benin HbS originated around Benin but expanded in the Sahara from which it diffused to other parts of North Africa and eventually made its way across the Mediterranean. This is even confirmed by its presence in predynastic Egyptian mummies! Which is not surprising considering that much of predynatic Egypt's population and culture come from the Sahara.
The map you provide seems to not answer my question. E3a is present at 9% in Palestine but where's the HbS? Also, seems that this simply jumped right past East Africa?
quote:recent where? Ask Mystery or Rasol, or better yet do research on your own.
How recent is HbS anyways?

quote:Yes, but in extremely low frequencies.
Originally posted by Djehuti:
^ But is not E3a also found among the Lower Nile Valley populations of Sudan and Egypt?
![[Big Grin]](biggrin.gif)
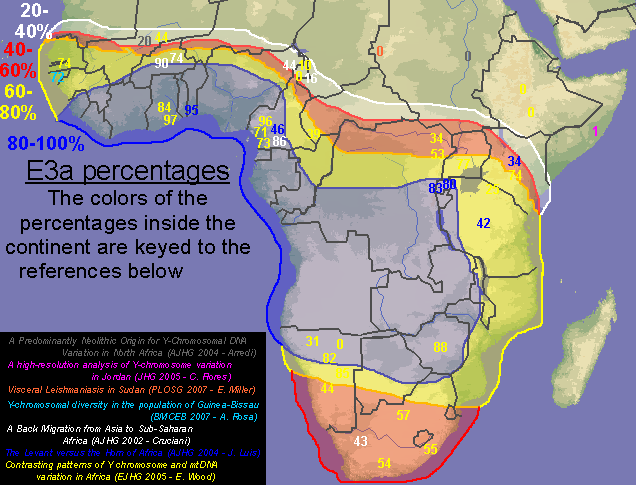
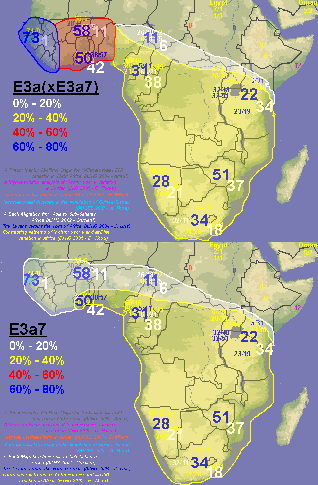
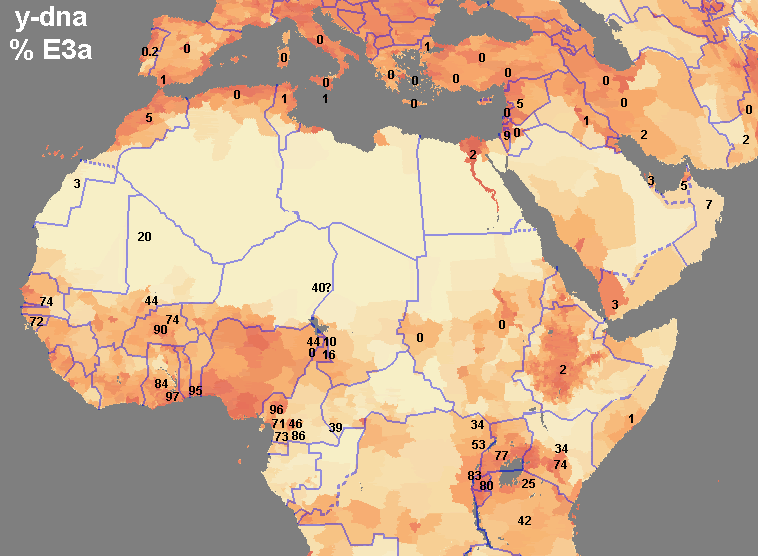
quote:I disagree, my map is clearly more detailed and with 10 times as much data as that one, plus it spans all of west Asia. Don't let those lines on the map you posted confuse you, that map was generated with only 10 or 12 data points, you can even distinguish them on the map, while I made mine with 100+ sampled populations. Many of the numbers on my map are the average of 2, 3 or 5 sampled populations that were near to each other.
Originally posted by Mackandal:
argiedude, here is a much better map of E3a distribution from a published study
quote:Evergreen Writes:
Originally posted by Charlie Bass:
quote:Yes, but in extremely low frequencies.
Originally posted by Djehuti:
^ But is not E3a also found among the Lower Nile Valley populations of Sudan and Egypt?
quote:On a number of studies, not just one. Some people are confusing haplotype IV with E3a which might be the problem.
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by Charlie Bass:
quote:Yes, but in extremely low frequencies.
Originally posted by Djehuti:
^ But is not E3a also found among the Lower Nile Valley populations of Sudan and Egypt?
What study are you using as a basis to this claim?
quote:Evergreen Writes:
Originally posted by Charlie Bass:
quote:On a number of studies, not just one. Some people are confusing haplotype IV with E3a which might be the problem.
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by Charlie Bass:
quote:Yes, but in extremely low frequencies.
Originally posted by Djehuti:
^ But is not E3a also found among the Lower Nile Valley populations of Sudan and Egypt?
What study are you using as a basis to this claim?
quote:And I hope you've learnt from it.
Originally posted by osiriun:
Thanks you for providing you own information and answering you own question. Wheww.
quote:Trivial. The fact of the matter is that the Asian haplotype is almost nearly exclusively restricted to south Asia [Indian Subcontinent] and eastern Arabia. No doubt the very *little* detection in Syria or Jordan, are the result of movements from Arabia. The more important point that you should have taken away from my posts, is that either side of the Mediterranean is in the main, affected by the African derivative called the Benin haplotype. The Benin haplotype 'distribution patterns' and variants belies the accounting of it in these regions by slavery. And even more importantly, in my posts, is the fact that the only type that occurs in both Egypt and Europe, is this derivative, the African derivative.
Originally posted by osiriun:
The majority of Syrians and Jordanians had the Benin haplotype, and severe disease. However, one in three Syrians and one in five Jordanians had a milder disease, and the Saudi-Indian haplotype was identified.
quote:^A necessary question. Other studies like the following [a recap btw], have suggested otherwise, if we are to acknowledge IV as the haplotype associated with the M2 mutation:
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by Charlie Bass:
quote:Yes, but in extremely low frequencies.
Originally posted by Djehuti:
^ But is not E3a also found among the Lower Nile Valley populations of Sudan and Egypt? [/qb]
What study are you using as a basis to this claim?
quote:This brings me back to my initial point which you're trying to moot.
Originally posted by Mystery Solver:
quote:Trivial. The fact of the matter is that the Asian haplotype is almost nearly exclusively restricted to south Asia [Indian Subcontinent] and eastern Arabia. No doubt the very *little* detection in Syria or Jordan, are the result of movements from Arabia. The more important point that you should have taken away from my posts, is that either side of the Mediterranean is in the main, affected by the African derivative called the Benin haplotype. The Benin haplotype 'distribution patterns' and variants belies the accounting of it in these regions by slavery. And even more importantly, in my posts, is the fact that the only type that occurs in both Egypt and Europe, is this derivative, the African derivative.
Originally posted by osiriun:
The majority of Syrians and Jordanians had the Benin haplotype, and severe disease. However, one in three Syrians and one in five Jordanians had a milder disease, and the Saudi-Indian haplotype was identified.
quote:No reason not to take IV in the African context as being associated with E3a chromosomes, not to mention buttressing by its distribution pattern. And yes, E3a has *definitely* been detected in Egypt. If there is reason to deny IV association with E3a chromosomes, I welcome its presentation.
Originally posted by Charlie Bass:
Only if haplotype IV is truly E3a could we say E3a is found in Upper Egypt/Northern Sudan.
quote:Taking care is something to be encouraged, however the situation with haplotype V is distinct from that of IV. Both V and XI chromosomes fall into E3b chromosomes for the most part in the African context, whereas IV doesn't have that element to it. Since RFLP doesn't actually identify the binary markers in of itself, this is where confusion was brought about in the contextualization of V and XI; I've gone through this a few times here before.
Originally posted by Charlie Bass:
I'm careful because of the problem we have had with haplotype V in Ethiopia. Haplotype V was supposed to be very high in Ethiopia yet E-M81 isn't.
quote:Relevant reading: Haplotype V revisited
Originally posted by Mystery Solver:
I've gone through this a few times here before.
quote:This map I posted was based on information from a huge dataset. If you go to the study and look at the dataset itself you would see this. At any rate E3a is has its highest frequencies in West Africa, not amongst Bantus.
Originally posted by argiedude:
quote:I disagree, my map is clearly more detailed and with 10 times as much data as that one, plus it spans all of west Asia. Don't let those lines on the map you posted confuse you, that map was generated with only 10 or 12 data points, you can even distinguish them on the map, while I made mine with 100+ sampled populations. Many of the numbers on my map are the average of 2, 3 or 5 sampled populations that were near to each other.
Originally posted by Mackandal:
argiedude, here is a much better map of E3a distribution from a published study
quote:Well, you are looking at the Data; the question is whether you're willing understand what you see. If you acknowledge that E3a has been detected in Egypt, then it should come as no shock whenever you come across it anywhere.
Originally posted by Charlie Bass:
I never denied that E3a is indeed in Egypt, I've just never saw it in the frequencies that correspond to those of haplotype IV.
quote:A study which only takes into account lower Egypt, which makes it all the more interesting, since this is an area where one would expect least probability [relatively speaking of course] of its detection if we were dealing with a lineage that is supposed to be of low frequency. Simply put, if lower Egypt can show this much detection of E3a, one can only imagine how much more would be detected going up the Nile Valley.
Originally posted by Charlie Bass:
In this study
The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors
of Human Migrations
E3a appears in Egypt at a frequency of 2.8%
quote:Arredi et al.'s study primarily focuses on E3b and J lineages, not E3a. Their samples, as far as I know consist of candidates from two locations: 44 candidates from Mansoura and 29 from Luxor. Of these samples, 14 binary markers were identified in the 44 candidates from Mansoura, while the 37 identifications were done by tandem repeats, and likewise, of the 29 candidates, 9 were actually defined by binary makers while 27 were defined only by tandem repeats. It is also of interest that this study also shows E3a detection in the Ethiopian sample. Whereas in the Luis et al. sample, more diverse speaking groups in northern Egypt were sampled, namely Arabic and Tamazight speakers. So it is the question of sampling selection, sampling size, and ability to detect as much binary markers along with STR markers as possible.
Originally posted by Charlie Bass:
And in this study
A Predominantly Neolithic Origin for Y-Chromosomal DNA Variation in
North Africa
E3a appears to be absent or at least almost absent from Northern(lower) and Southern(Upper) Egypt and Sudan.
quote:Your suspicion to me is unfounded, because you yourself acknowledge that at least one of the studies you cited *explicitly* notes E3a detection in Egypt, and yet you are perplexed by its finding in another, wherein again relatively lower detection occurs in lower Egypt and relatively higher frequencies found in the Upper Nile. The distribution makes sense. What else is even telling, is that you provide no reason as to why IV could be anything but M2 affiliated chromosomes as Keita has observed; instead, you simply tell us that your *suspicions* are holding you back. I've provided more than a "suspicion" to demonstrate why there is no reason to doubt that IV associates with M2-associated chromosomes.
Originally posted by Charlie Bass:
Thats why I'm suspicious as of the correlation of haplotype IV with E3a.
quote:That its there isn't shocking to me, I simply stated that the frequency of E3a doesn't correspond to the frequencies of haplotype IV found in Lucotte et al's study If you think otherwise you need to produce evidence for it.
Originally posted by Mystery Solver:
[QB]quote:Well, you are looking at the Data; the question is whether you're willing understand what you see. If you acknowledge that E3a has been detected in Egypt, then it should come as no shock whenever you come across it anywhere.
Originally posted by Charlie Bass:
I never denied that E3a is indeed in Egypt, I've just never saw it in the frequencies that correspond to those of haplotype IV.
quote:You got me totally wrong, and what about the virtual absence of E3a in Sudan, yet Lucotte et al says higher frequencies of haplotype IV is supposed to be there? I have perfectly good reasons for having some suspicion about the correlation between haplotype IV and E3a.
Your suspicion to me is unfounded, because you yourself acknowledge that at least one of the studies you cited *explicitly* notes E3a detection in Egypt, and yet you are perplexed by its finding in another, wherein again relatively lower detection occurs in lower Egypt and relatively higher frequencies found in the Upper Nile. The distribution makes sense. What else is even telling, is that you provide no reason as to why IV could be anything but M2 affiliated chromosomes as Keita has observed; instead, you simply tell us that your *suspicions* are holding you back. I've provided more than a "suspicion" to demonstrate why there is no reason to doubt that IV associates with M2-associated chromosomes.
quote:And what specific lineages could have possibly been lost?
Originally posted by Evergreen:
Evergreen Writes:
It is probable that E3a/M2 was much higher in Lower Egypt pre Late Dynastic times. Many of the indigenous African lineages were swamped due to non-African infiltration into NE Africa at this time.
quote:Evergreen Writes:
Originally posted by Charlie Bass:
quote:And what specific lineages could have possibly been lost?
Originally posted by Evergreen:
Evergreen Writes:
It is probable that E3a/M2 was much higher in Lower Egypt pre Late Dynastic times. Many of the indigenous African lineages were swamped due to non-African infiltration into NE Africa at this time.
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]You got me totally wrong, and what about the virtual absence of E3a in Sudan
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]That its there isn't shocking to me, I simply stated that the frequency of E3a doesn't correspond to the frequencies of haplotype IV found in Lucotte et al's study If you think otherwise you need to produce evidence for it.
quote:That may certainly be true, although it looks as if E-M81 and E-M78 look to be the predominant lineages that may have ben in abundance before non-African infiltration.
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by Charlie Bass:
quote:And what specific lineages could have possibly been lost?
Originally posted by Evergreen:
Evergreen Writes:
It is probable that E3a/M2 was much higher in Lower Egypt pre Late Dynastic times. Many of the indigenous African lineages were swamped due to non-African infiltration into NE Africa at this time.
Obviously E3a.
quote:Thats why I said *VIRTUAL* absence, there is some E3a in Sudan, but the frequencies don't correlate with the high frequencies of haplotype IV that were observed in Lucotte et al's study. If people of West African ancestry moved there and they have, there would be some E3a there, but this wouldn't have any bearing on E3a in prehistoric times.
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]You got me totally wrong, and what about the virtual absence of E3a in Sudan
That's seems to be a far-fetched claim. Heck, there are people of recent West African origin in the Sudan. How could E3a possibly be "virtually absent". Please provide your source.
quote:From what I've read on haplotype IV, it does in fact appear to correlate to E3a in West Africa, but I cannot say the same for Upper Egypt and Northern Sudan regarding this.
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]That its there isn't shocking to me, I simply stated that the frequency of E3a doesn't correspond to the frequencies of haplotype IV found in Lucotte et al's study If you think otherwise you need to produce evidence for it.
What frequency does IV correspond to based upon your research?
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]That may certainly be true, although it looks as if E-M81 and E-M78 look to be the predominant lineages that may have ben in abundance before non-African infiltration.
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]Thats why I said *VIRTUAL* absence, there is some E3a in Sudan, but the frequencies don't correlate with the high frequencies of haplotype IV that were observed in Lucotte et al's study.
quote:Evergreen Writes:
Originally posted by Charlie Bass: [QUOTE]If people of West African ancestry moved there and they have, there would be some E3a there, but this wouldn't have any bearing on E3a in prehistoric times.
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]From what I've read on haplotype IV, it does in fact appear to correlate to E3a in West Africa, but I cannot say the same for Upper Egypt and Northern Sudan regarding this.
quote:True
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]Thats why I said *VIRTUAL* absence, there is some E3a in Sudan, but the frequencies don't correlate with the high frequencies of haplotype IV that were observed in Lucotte et al's study. [/qb]
This could be due to sampling size and variation in the regions tested, correct?
quote:True.
Originally posted by Charlie Bass:quote:Evergreen Writes:
If people of West African ancestry moved there and they have, there would be some E3a there, but this wouldn't have any bearing on E3a in prehistoric times.
You are correct. However, we know that Saharans moved into the Nile and that the same source population feed West Africa. A common gene pool is to be expected, correct? [/QB]
quote:I've already stated it. I just haven't seen many studies that show a frequency of E3a in Upper Egypt/Sudan that parallels the frequency of Haplotype IV in these same areas. Perhaps haplotype IV could be affiliated with more than just E3a, depending upon the population.
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]From what I've read on haplotype IV, it does in fact appear to correlate to E3a in West Africa, but I cannot say the same for Upper Egypt and Northern Sudan regarding this.
Ok, but you have alluded my original question:
What frequency does IV in Egypt/Sudan correspond to based upon **YOUR** research?
quote:I had already seen this map a few months ago. It's from the 2007 study of Guinea-Bissau. After taking a 2nd look at the data accompanying their map, I was surprised at how many samples they used, since, I'm sorry to say, their map seems to be "choppy". But all of those samples I already knew about and included them in my map. It was good to know, reading that list, that I have done a thorough job investigating the scientific reports on E3a in Africa. I didn't miss a single one. And I included a few more that they didn't, such as the 200+ samples of people originating from Darfur (no E3a was found). And like I said previously, my map also includes E3a outside of Africa, which is extremely interesting in itself.
Originally posted by Charlie Bass:
quote:This map I posted was based on information from a huge dataset. If you go to the study and look at the dataset itself you would see this. At any rate E3a is has its highest frequencies in West Africa, not amongst Bantus.
Originally posted by argiedude:
quote:I disagree, my map is clearly more detailed and with 10 times as much data as that one, plus it spans all of west Asia. Don't let those lines on the map you posted confuse you, that map was generated with only 10 or 12 data points, you can even distinguish them on the map, while I made mine with 100+ sampled populations. Many of the numbers on my map are the average of 2, 3 or 5 sampled populations that were near to each other.
Originally posted by Mackandal:
argiedude, here is a much better map of E3a distribution from a published study
quote:I know people are not static, I just hinted that maybe E-M81 and E-M78 were the most abundant haplogroups in Egypt. I could as well be wrong and yes some R* lineages could have originated in NE Africa.
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]That may certainly be true, although it looks as if E-M81 and E-M78 look to be the predominant lineages that may have ben in abundance before non-African infiltration.
This is not true. NE African bio-history is not static. Various lineages would have been present in pre Late Dynastic Egypt. This would vary based upon the region and the time frame. Some lineages that existed may now be extinct for that matter. Some lineages such as y-chromsome lineage R* may be indigenous to NE Africa as well.
quote:Evergreen Writes:
Originally posted by Charlie Bass:
[QUOTE]I've already stated it. I just haven't seen many studies that show a frequency of E3a in Upper Egypt/Sudan that parallels the frequency of Haplotype IV in these same areas. Perhaps haplotype IV could be affiliated with more than just E3a, depending upon the population.
quote:Evergreen Writes:
Originally posted by argiedude:
[QUOTE]I included a few more that they didn't, such as the 200+ samples of people originating from Darfur (no E3a was found).
quote:http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17500593
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by argiedude:
[QUOTE]I included a few more that they didn't, such as the 200+ samples of people originating from Darfur (no E3a was found).
What is your peer-reviewed source for this claim?
quote:Makes no sense. You say that E3a doesn‘t correspond to the frequencies of haplotype 4, even though an extract from a study was cited *explicitly* stating that the former does associate with the latter.
Originally posted by Charlie Bass:
That its there isn't shocking to me, I simply stated that the frequency of E3a doesn't correspond to the frequencies of haplotype IV found in Lucotte et al's study If you think otherwise you need to produce evidence for it.
quote:Who said it is absent, i.e. if by Sudan, you are referring to "lower Nubia"? Not according to data used by Lucotte et al. or Keita, that is for sure; in fact, that data says otherwise.
Originally posted by Charlie Bass:
You got me totally wrong, and what about the virtual absence of E3a in Sudan
quote:It makes sense for higher frequencies to be in the region straddling Egypt and northern Sudan, since it is naturally closer to sub-Saharan Africa than the coastal north regions. But this is just a distraction; if you have evidence that haplotype IV doesn't correspond to M2 bearing chromosomes as already cited, then produce it.
Originally posted by Charlie Bass:
yet Lucotte et al says higher frequencies of haplotype IV is supposed to be there?
quote:But you produce no substantive merit to back it up.
Originally posted by Charlie Bass:
I have perfectly good reasons for having some suspicion about the correlation between haplotype IV and E3a.
code:1 - Lucotte and Mercier (2003a)
Population (n) Haplotypes and percentages
IV V XI VII VIII XII XV
Falasha (38) 0.0 60.5 26.3 0.0 0.0 0.0 0.0
Ethiopians [non Falasha](104) 0.0 40.4 25.9 0.0 23.1 0.0 0.0
Berbers (74) 1.4 68.9 2.8 1.4 6.8 4.1 0.0
(Morocco)
“Sephardic” Jews (381) 8.4 18.6 6.8 19.9 34.1 4.2 2.1
“Oriental” Jews(56) 1.8 8.9 0.0 7.1 78.6 0.0 1.8
“Near Eastern” (27) 0.0 7.4 0.0 7.4 85.1 0.0 0.0
Askenazic Jews(256) 0.0 3.1 15.2* 22.7 24.6 9.0 10.9
code:6 - Lucotte et al. (1996)country (n) Haplotypes and percentages
IV V XI VII VIII XII XV
Egypt(274) 13.9 39.4 18.9 6.6 7.3 2.2 5.5
Lebanon(54) 3.7 16.7 7.4 20.4 31.5 5.6 1.9
Palestine(69) 1.4 15.9 5.8 13.0 46.4 0.0 4.3
Iraq(139) 1.4 7.2 6.4* 20.1 36.0 1.4 0.7
Egypt(52) 7.7 40.4 21.2 9.6 7.7 3.8 1.9
Libya (38) 7.9 44.7 10.5 0.0 5.3 13.2 0.0
Algeria (141) 8.5 56.7 5.0 1.4 7.1 4.2 5.0
Tunisia (73) 0.0 53.4 5.5 4.1 2.7 26.0 2.7
Morocco (102) 0.98 57.8 8.8 4.9 7.8 0.98 10.8
Mauretania (25) 8.0 44.0 8.0 0.0 4.0 0.0 0.0
Suprasah(composite)(505)
4.4 55.0 7.7 3.2 6.3 7.1 4.2
Ethiopia(composite)(142) 0.0 45.8 26.1 0.0 16.9 0.0 0.0
quote:To understand how Al Zahery et al. were able to link RFLP haplotypes to SNP markers on the chromosomes under study, they identified the former by using TaqI restriction enzyme digests and the latter with the likes of PCR and DHPLC analysis; essentially this sort of approach to testing for both RFLP markers and binary markers has been exemplified in the following link - we discussed the methods utilized in relative detail, in the sorting out of RFLP markers into respective sub-clades: Haplotype V revisited
Originally posted by rasol:
However this is based on the assumption that TaqI can be reliably mapped to Y chromosome.
I don't understand the relationship well enough to comment on that.
quote:Already provided. Please cater to my outstanding requests.
Originally posted by Charlie Bass:
Supercar, please post a study that shows substantial frequencies of E3a in Sudan and Upper Egypt that correspond to the frequencies of haplotype IV in Lucotte et al's study.
quote:Evergreen Writes:
Originally posted by Mystery Solver:
[QUOTE]Haplotype IV, designating the M2/PN1 subclade, as noted, is found in high frequency in west, central, and sub-equatorial Africa in speakers of Niger-Congo—which may have a special relationship with Nilosaharan—spoken by Nubians; together they might form a superphylum called
Kongo-Saharan or Niger-Saharan (see Gregersen 1972, Blench 1995), but this is not fully supported.

quote:Evergreen Writes:
Originally posted by Charlie Bass:
A few of trhe studies I've saw that have tested Nilo-Saharans don't show a high frequency of E3a. The Maasai and datoga both show high frequencies of M35 and A3b2 and relatively lower frequencies of E3a which *COULD* be due to recent mixture with Bantu speakers.
quote:Yes to all, and I'll like to add that each study has to be carefully reviewed on its methodological merits, which includes regions sampled, sample size, *target* markers of the authors and actual markers found, regional and ethnic diversity of samples.
Originally posted by Evergreen:
Evergreen Writes:
Key takeaways -
A. The research community investigating the phylogenetic history of haplogroup E has a history of working with patterns of conscious and subconscious bias.
B. Little research on haplogroup E3a has been conducted.
C. Sample sizes and samples of a wide variety of populations in Chad, Sudan, Egypt, Niger, etc have not been conducted.
D. When samples are taken inappropriate models based upon stereotype and bias have been constructed.
E. Wide samples of Nilo-Saharan have not been taken.
F. Archaeology and linguistics indicate Nilo-Saharan inputs into Neolithic and Pre-Dynastic Egypt (Southern Egypt and Northern Sudan).
G. Nilo-Saharan and Niger-Congo languages seem to have a deep time-depth relationship and this is congruent with a shared pre-LGM heritage. This would indicate a common E3a ancestry for these populations. This is consistent with what we know of haplogroup E distributions among Senegalese populations.
quote:You do realize what RFLP haplotype IV corresponds to, right?
Originally posted by Charlie Bass:
Same for haplotype IV, its absent in the Central African Republic and in Pygmies according to one study I read.
quote:Evergreen Writes:
Originally posted by argiedude:

This map shows the distribution of E3a and the population density.
quote:You're probably wasting your time in pointing out any shortcomings; I've tried, and it proved to be futile.
Originally posted by Evergreen:
Evergreen Writes:
Your map needs work. Does it not strike you as odd that E3a approaches frequencies near 10% in Palestine and 0% in Upper Egypt according to your graphic.
quote:Evergreen Writes:
Originally posted by Mystery Solver:
quote:^A necessary question. Other studies like the following [a recap btw], have suggested otherwise, if we are to acknowledge IV as the haplotype associated with the M2 mutation:
Originally posted by Evergreen:
quote:Evergreen Writes:
Originally posted by Charlie Bass:
quote:Yes, but in extremely low frequencies.
Originally posted by Djehuti:
^ But is not E3a also found among the Lower Nile Valley populations of Sudan and Egypt?
What study are you using as a basis to this claim?
Of note are the frequencies of the aforementioned haplotypes; V, XI, and IV:
Lower Egypt (n=162); V=51.9%, XI=11.7%, and IV=1.2%.
Upper Egypt (n=66); V=24.2%, XI=28.8%, and IV=27.3%.
Lower Nubia (n=46); V=17.4%, XI=30.4%, and IV=39.1%.
The primary haplotypes found in Europe, XII and XV, according to Keita, could have likely entered into the country via Greco-Roman invasions, are found in Lower Egypt at 3.7% and 6.8% respectively. Not surprising, the frequencies of these two are lower in Upper egypt, with XII not represented, while XV is at 6.1%. They are absent in Lower Nubia. These results are according to Lucotte and Mercier's findings, 2003
Details discussed here: http://thenile.phpbb-host.com/phpbb/ftopic152-15.php [/QB]
quote:Evergreen Writes:
Originally posted by Evergreen:
[QUOTE]
Evergreen Writes:
Hence we see a smooth grade of haplogroup E3a spreading out of NE Africa during the Early Bronze Age.
Syria = ~ 5%
Palestine = ~ 10%
Lower Egypt = ~ 2%**
Upper Egypt = ~ 28%
Lower Nubia = ~ 39%
** Lower Egypt may be an outlier due to the infiltration of large numbers of non-indigenous Africans post-Late Kingdom Dynastic era.
Haplogroup E3a may have spread from the Central Sahara to the Nile Valley during the pre-early Neolithic phase. There is further evidence of the spread of Saharan cultural elements such as bi-facial arrowheads and pottery into the early neolithic Faiyum and southern Levant. Further itterations of immigration may have followed with the early Dynastic and Middle Bronze Age Egyptian colonial activity in "Canaan".
West Africa and the Central Sahara in turn may be connected via the movement of early holocene, pottery and bi-facial arrowhead (Ounan points) using populations from West African regions such as Ounjougou Mali into the Central Sahara. Hence, West Africa is a likely population source of the early Holocene Central Sahara and by default early neolithic base population of ancient Egypt.
quote:
Originally posted by Evergreen:
[QUOTE]
Sub-populations within the major European and African derived haplogroups R1b3 and E3a are differentiated by previously phylogenetically undefined Y-SNPs.
Sims et. al.
Single nucleotide polymorphisms on the Y chromosome (Y-SNPs) have been widely used in the study of human migration patterns and evolution. Potential forensic applications of Y-SNPs include their use in predicting the ethnogeographic origin of the donor of a crime scene sample, or exclusion of suspects of sexual assaults (the evidence of which often comprises male/female mixtures and may involve multiple perpetrators), paternity testing, and identification of non- and half-siblings. In this study, we used a population of 118 African- and 125 European-Americans to evaluate 12 previously phylogenetically undefined Y-SNPs for their ability to further differentiate individuals who belong to the major African (E3a)- and European (R1b3, I)-derived haplogroups. Ten of these markers define seven new sub-clades (equivalent to E3a7a, E3a8, E3a8a, E3a8a1, R1b3h, R1b3i, and R1b3i1 using the Y Chromosome Consortium nomenclature) within haplogroups E and R. Interestingly, during the course of this study we evaluated M222, a sub-R1b3 marker rarely used, and found that this sub-haplogroup in effect defines the Y-STR Irish Modal Haplotype (IMH). The new bi-allelic markers described here are expected to find application in human evolutionary studies and forensic genetics.